CM 793 Presentation Date: November 11, 2021 @ 2:00 PM in Pathology 103
Title: “Identifying HopBA1 recognition requirements”
Advisor: Marc Nishimura

CM 793 Presentation Date: November 11, 2021 @ 2:00 PM in Pathology 103
Title: “Identifying HopBA1 recognition requirements”
Advisor: Marc Nishimura
ABSTRACT:
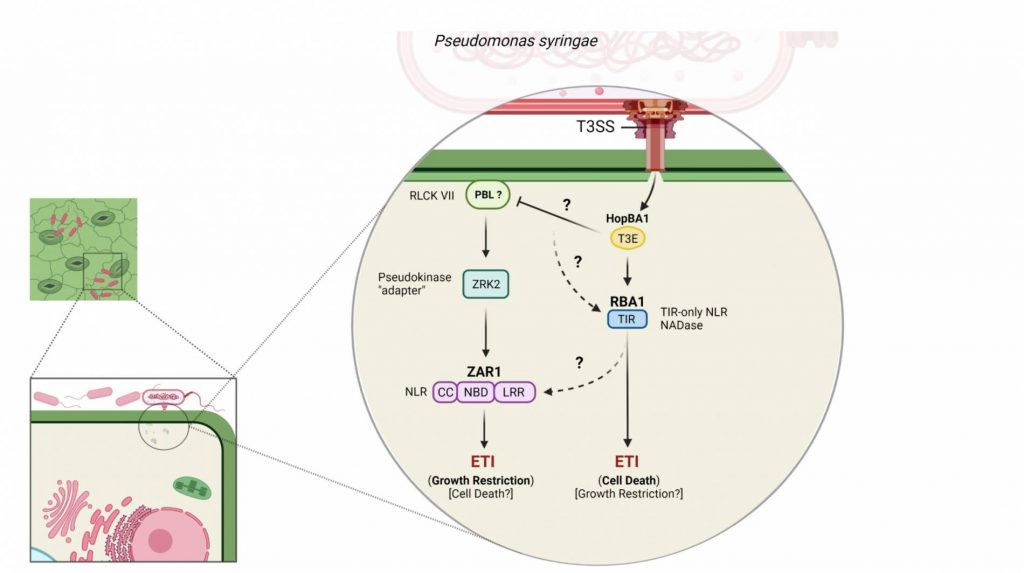
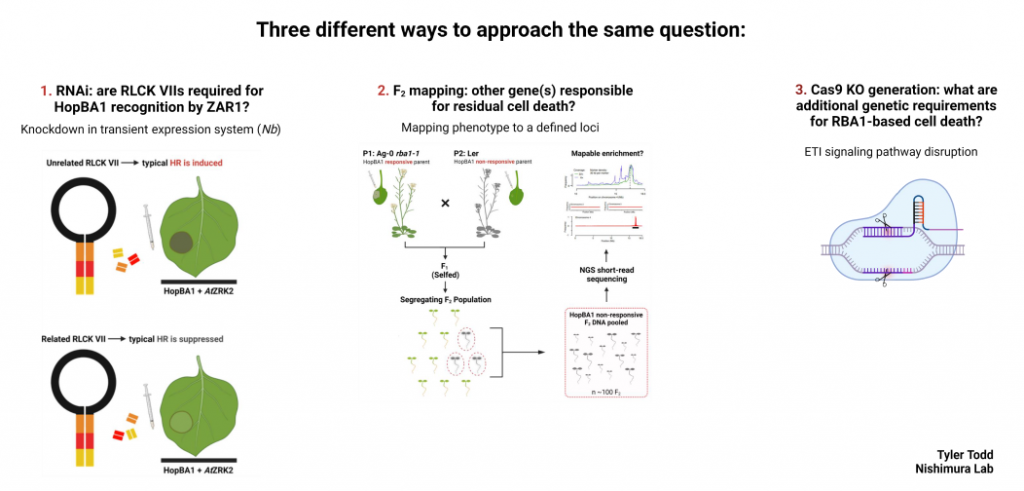
Plant pathogens have evolved numerous methods to evade detection by the plant immune system. One method is by secreting small proteins, known as effectors, into the cytoplasm of the cell. Once inside, effectors may modify host proteins, transcriptional activity, etc., in a way that allows proliferation or survival vs. detection and the initiation of basal defense responses. In a step of counter-evolution, adapted plant species respond to this challenge with a suite of defense responses generally referred to as effector triggered immunity (ETI). Specifically, we study a type III effector known as HopBA1, which Pseudomonas syringae injects into plants using its type III secretion system. Previously, a non-canonical immune receptor, RBA1, was identified for its role in HopBA1-triggered cell death. Later, another immune receptor, ZAR1, was identified as recognizing HopBA1. ZAR1 recognition, seemingly, involves an entirely different pathway and mechanism of ETI. This project ultimately seeks to resolve standing questions on HopBA1 activity within the plant cell, as well as its perception by the plant immune system. This probe is largely a genetic one, utilizing RNAi to silence candidate HopBA1-target proteins, Cas9 to knockout candidate immune signaling genes, and a more classical F2 population to potentially identify and map additional genetic requirements
for HopBA1-triggered cell death.